Defining
Evolution
And
its Boundaries
Sean
D. Pitman M.D.
©
September 2006
The
theory of evolution is based on a
very simple and elegant idea. The idea is that all living things have a common
ancestor. The differences between living things that exist today are thought to
be the result of "common
decent with modification" or slight changes over time that have simply
added up over many millions of generations to produce the remarkable variety
that we see today. These modifications are the result of mindless
non-directed random genetic mutations combined with natural selection. Natural selection is able to
select between these randomly produced genetic sequences in such a way that
those sequences with the greatest attached beneficial function provide their
host the the greatest reproductive advantage. In this way, natural
selection, while still mindless or non-intentional, is the non-random part of
evolution. 3,4,5,6
If
the amazing diversity of life forms on this planet arose from the evolutionary
potential of a common ancestor life form, the assumption can be made that all or
nearly all life forms living today have the same potential for future diversity.
If true, this mindless force is a very creative force.
But, how does this mindless force work?
Most
will agree that if living things change over time, they change because their
D.N.A. (deoxyribonucleic acid) changed. The
information contained in the DNA is called the “genotype.â€Â
The expression of this information in the physical form of the creature
is called the “phenotype.†1, 2
DNA is very much like the paper that the blueprint for a house is written
down on. This blueprint is
equivalent to the genotype. The
actual house, once it is built, is equivalent to the phenotype.
The phenotype changes only if the blueprint changes.
 The
theory of evolution proposes that all the various blueprints of living things
are descendents of a single common ancestor blueprint.
The diversity of blueprints that exist today is simply the result of
variations on a single theme. If
true, the power of the mindless evolutionary force of nature is truly
astounding. The sheer creativity
and magnificent diversity of nature is enough to make even the most cynical
stand in silent awe. If humankind
could harness this power and speed it up with the aid of our intelligent minds,
the implications for advancement seem unlimited.
The
theory of evolution proposes that all the various blueprints of living things
are descendents of a single common ancestor blueprint.
The diversity of blueprints that exist today is simply the result of
variations on a single theme. If
true, the power of the mindless evolutionary force of nature is truly
astounding. The sheer creativity
and magnificent diversity of nature is enough to make even the most cynical
stand in silent awe. If humankind
could harness this power and speed it up with the aid of our intelligent minds,
the implications for advancement seem unlimited.
How
then does this mindless evolution work? How
does the equivalent of a blueprint for a house change over time to code for
phenotypic structures as diverse as an automobile, a battleship, a skyscraper,
or a screwdriver? Obviously, at least this
degree of diversity is seen in living things in the forms of creatures like bacteria, oak trees, and
elephants. In considering this question perhaps we should begin
with Darwin and what he saw. (Back to Top)
Charles
Darwin
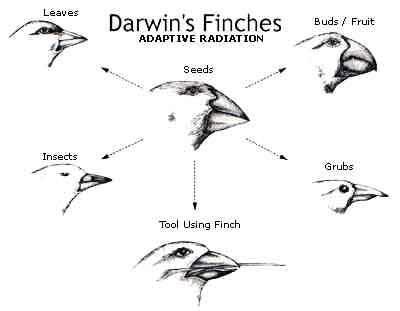
Charles
Darwin (1809-1882) came up with his famous version of the theory of evolution
after observing some very interesting differences, such as the variation in the size
and shape of finch beaks on the Galapagos Islands.
Many other similar changes have also been observed and carefully documented.
Certainly these are “changes†and as changes many would call them
evolutionary changes. If the theory
of evolution is defined as any and every phenotypic change that occurs from
parents to offspring, then it might be perfectly fine to say that finches are
demonstrating evolution in real time. But,
are they demonstrating genotypic evolution?
Has the blueprint changed in an informationally unique way?
Is it possible to have change in phenotype without a change in genotype? In
other words, do the phenotypic changes in the finches that Darwin observed require
new meaningful genetic information that the ancestor finches never had? (Back
to Top)
Gregor
Mendel
 Darwin
was unable to answer this question although the answer was in fact available in
his own day. Gregor
Mendel (1822-1884), the father of genetics, came up with the idea of
“alleles†or unchanging “traits†after studying pea plants. 3,7 What
he discovered is that certain phenotypic traits, such a pea color, texture,
shape, and a number of other traits, are passed on by unchanging genotypic
alleles. Different combinations of
these alleles result in different allelic expressions in the phenotype.
Darwin
was unable to answer this question although the answer was in fact available in
his own day. Gregor
Mendel (1822-1884), the father of genetics, came up with the idea of
“alleles†or unchanging “traits†after studying pea plants. 3,7 What
he discovered is that certain phenotypic traits, such a pea color, texture,
shape, and a number of other traits, are passed on by unchanging genotypic
alleles. Different combinations of
these alleles result in different allelic expressions in the phenotype.
For
example, lets say that a house needs colored carpet.
Colored carpet is a “trait†or characteristic of the house listed in
the blueprint. The blueprint of this
particular house is interesting however in that it is a double blueprint.
There are two pieces of paper that code for every aspect of the house. The
two blueprints are identical as far as the traits that they code for (ie:
colored carpet), but they are different as far as the trait variations are
concerned (ie: red, yellow, green, blue or white carpet). If one blueprint
coded for green carpet and the other coded for white carpet, what color would
the carpet be? It will be the “dominant†color.
Mendel
found that allelic traits could be either dominant or recessive (We now know
that they can also be co-dominant, incompletely dominant, additive,
multiplicative etc.) This is made
possible because of the fact that many traits have at least two alleles, or two
separate codes on two blueprint copies, that code for the same trait.
If both alleles are the same, then the expressed phenotype will match
both alleles. If the two alleles
are different, then the phenotypic expression will match the dominant allele.
Each allele is inherited unchanged, one from each parent.
During the process of sexual reproduction, the alleles coding for the
same trait trade places with each other randomly (from one blueprint copy to the
same place on the other blueprint copy) so that the next generation will be
uniquely different from the current generation in their phenotypic expression of
the same alleles. This is why
siblings from the same parents never look exactly alike unless they are twins
arising from the same fertilized egg. Siblings
can in fact look very different from each other and even their parents.
One may have a big nose and the other a small nose.
Similarly, a finch may have a bigger or smaller beak than its siblings.
Such phenotypic changes do indeed occur, but they need not be based in any
change of the common "gene pool" of options. 2
(Back to Top)
Breeding
via Human Selection
 The
variation in allelic expression can be quite dramatic.
It is responsible for the majority of changes seen in animal breeding.
For example, the main differences between a Chihuahua and a Great Dane
are primarily the result of trait selection where desired traits contained by a
common gene pool were gathered together over a few generations into one animal.
Most of traits themselves already existed, fully formed, in the common ancestral
gene pool of dogs. Thus, neither of
these breeds has “evolved†much of anything that their common ancestors did
not already have in their common gene pool of options.
The
variation in allelic expression can be quite dramatic.
It is responsible for the majority of changes seen in animal breeding.
For example, the main differences between a Chihuahua and a Great Dane
are primarily the result of trait selection where desired traits contained by a
common gene pool were gathered together over a few generations into one animal.
Most of traits themselves already existed, fully formed, in the common ancestral
gene pool of dogs. Thus, neither of
these breeds has “evolved†much of anything that their common ancestors did
not already have in their common gene pool of options.
The
ability for great phenotypic variation is obvious, but there are clear limits to
this variation. Using genetic
recombination alone, a dog cannot be anything but a dog.
A dog can never be changed, via genetic recombination alone, into a cat
or a chicken or anything else. Why?
Because cats, dogs, and chickens are made from different blueprints that
code for different trait types and trait options that are not contained by the
gene pools of the others. Also, traits that may be similar may not
necessarily be located at the same relative positions on their respective
genetic blueprints.
Again,
using the house blueprint analogy as an example, one house might have a
blueprint that codes for carpet at the bottom right-hand corner of the page.
Another house might have a blueprint that codes for an electric garage door
opener at this location and has no code for carpet at all. Also, the first
house might not have a code for a garage much less an electric garage door
opener. Neither one of the blueprints for one house will match with the
options or order of options for the other house.
So,
what does this mean?
It means that the blueprints for the different houses in this case cannot talk
to each other, mix and match, or "recombine" their collective
information in any functional way. They cannot
"interbreed" so to speak to produce viable "offspring".
Only blueprints that have the same setup and "trait" types can
exchange equivalent information with each other. It is impossible then for
blueprints that do not have a position for a garage code to trade equivalent
information that results in the formation of a garage. The same is true
for different animals such as dogs, chickens and cats. They cannot breed
with each other, nor can they be bred to look like each other, using genetic
recombination alone.
Although
genetic
recombination can and does result in some very dramatic phenotypic changes for
these creatures, this process is limited by the edges of a large
but finite pool of options. Such a
gene pool remains fixed while various creatures within the gene pool give
phenotypic expression to various aspects of this genotypic pool of options.
The pool provides the means for huge phenotypic variation or “changeâ€Â
but the genotypic pool itself may not change from one generation to the next. 8
The changing phenotypic creature is nothing more than a partial
reflection of a non-changing or "static" genotypic pool. Thus,
it is the genotypic pool and not the phenotypic creature that "real" evolution must act
on.
But, if Darwin’s finch
beaks are not examples of gene pool evolution is there anything that is?
Is there any creature that has unique traits that its ancestors did not
have in their gene pool? (Back
to Top)
Random
Mutations
This
is where mutations come into play. Genetic mutations are relatively rare
random changes that occur in a creature's genotypic blueprint that were not in
the blueprints of that creature’s parents.
There are many different types of mutations.
There are point mutations where just one letter is changed in the wording
of the genetic blueprint. There are
translocation mutations where a section of the blueprint is cut out and pasted
in another place on the blueprint. There
are inversion mutations where a section of the blueprint is cut out and turned
upside down and pasted back in the same place.
There are duplication mutations where a section of the blueprint is
copied and then pasted in another place. The
list goes on and on, but basically the mutated blueprint has genes/alleles or
genetic sequences that were not in the blueprints of either one of the parent
blueprints. 1,2,4
(Back to Top)
Functional
vs. Neutral Mutations
As
would seem intuitive, most functional mutations are harmful and may even
be lethal. Fortunately though, most
mutations are not functional. Most mutations are silent or
"neutral" and result in no detectable phenotypic change. Very
rarely, some mutations are “beneficial.â€Â
The ratio of beneficial vs. detrimental mutations is on the order of 1 in 1,000
for certain types of functions (discussed in more detail below).
Common examples of beneficial mutations are those that give bacteria antibiotic
resistance or those that cause sickle cell anemia in people who live where
malaria is prevalent. But how,
exactly, do beneficial mutations achieve their benefits? (Back
to Top)
Antibiotic
Resistance
In
the case of bacterial
antibiotic resistance, certain mutations actually do result in the fairly
sudden creation of new as well as beneficial functions that the
"parent" bacteria did not have in their collective gene pool of
options.
Now,
it should be noted that bacteria are not like dogs, cats, and
chickens, or anything else that uses sex or genetic recombination as a means of
reproduction. Bacteria reproduce themselves by a relatively simple method
of cell division. In other words, all of the offspring of a given
bacterium will be identical with itself as well as with each other. They
are basically clones of each other. Because of this, there are no trait
options to choose from. There is only one copy of the blueprint instead of
the two copies used in sexual recombination. So, there is only one option for
each bacterial "trait." There is no gene "pool" of options
for each trait - so to speak. Of course, many types of bacteria can in fact
laterally exchange genetic information via
plasmids and the like. But, as a general rule of thumb, bacteria do not
undergo genetic recombination. So, for all practical purposes, all bacteria within a given
isolated population are
identical except if a mutational change occurs. Such mutations,
when they do occur, are passed on to all subsequent offspring.
So,
back to the notion of beneficial mutations. They do happen in bacterial
populations all the time. For example,
penicillin resistance is not always gained by the production of the famous
ß-lactamase
enzyme "penicillinase." There are several other ways that
bacteria become resistant to penicillin. A notable example occurs in Streptococcus
pneumoniae bacteria. ß-lactamase
have never been identified in S. pneumoniae and yet they are capable of
penicillin antibiotic resistance due to modification of their penicillin binding
proteins (PBPs). Since PBPs are the natural target of penicillin, many
different point mutations within this target are capable of interfering with the
target-antibiotic interaction. It is this interference that results in
penicillin resistance. And, importantly, this resistance, combined
with what are called "compensatory
mutations", can be achieved without any significant loss of any other
functional system within the bacterium. So, the argument that all
mutations end up producing at least some detectably harmful effect to gain a
beneficial effect simply isn't true.
Other
antibiotics require a specialized transport protein to bring them into the
bacterium. Again, many different point mutations can interfere with the
ability of the transport protein to interact properly with the antibiotic.
This interference results in resistance to this particular antibiotic. Again,
this interference can be gained without any significant functional loss of
the mutant bacteria relative to their peers.
Other
bacteria already make more complex antibiotic enzymes, such as the penicillinase
enzyme. Such enzymes do not evolve in previously susceptible
bacterial populations. There is not a single documented case where the
penicillinase enzyme code has been observed to evolve in a population where it
wasn't already there. The coded sequence or "gene" needed to
produce the penicillinase enzyme was already there or it was gained via lateral
transfer from other bacteria who already had this code (often via plasmids).
The problem is that this coded sequence is usually regulated so that the
penicillinase enzyme is not produced in sufficient enough quantities to protect
the bacterium from high levels of the penicillin antibiotic. Several
different mutations are capable of blocking or interfering with the suppression of penicillinase
production so that much greater quantities can be made, which results in
enhanced penicillin resistance.
Similarly,
Mycobacterium tuberculosis, the cause
of the tuberculosis disease, produces an enzyme that (as well as its other
useful functions) changes the non-harmful antibiotic "isoniazid" into
its active and lethal form. The now
active isoniazid proceeds to kill the Mycobacterium. Several
different mutations are capable of interfering with the isoniazid-enzyme
interaction. And again, this interference results in Mycobacterial resistance
to isoniazid.
To
give another example, the 4-quinolone antibiotics attack the enzyme “DNA
gyrase†inside various bacteria. Again,
several different point mutations are capable of interfering with the gyrase-antibiotic
interaction.9
Perhaps
the most famous and oft quoted example of a beneficial mutation is the point mutation of the hemoglobin
molecule that is seen in people affected by a condition known as sickle cell anemia.
This single point mutation decreases the effective oxygen carrying capacity of
the hemoglobin molecule.
It still carries oxygen, just not as well.
Now, it just so happens that the malarial parasite needs a high oxygen concentration to survive and so
cannot survive in blood with the sickle cell mutation.10 Those
people who have only one of their two blueprint DNA copies affected by this
mutation do not have significant anemia, but their blood still doesn't carry
oxygen well enough to support the malarial parasite. So, they are
resistant to malaria while at the same time having little problem with the
hemoglobin mutation. However, those unfortunate individuals who end up
with a double mutation, known as a "homozygous" condition, have a
severe problem with their hemoglobin molecules crystallizing under low oxygen
tension. This crystallization effect dramatically distorts the red blood cells
and they no longer fit very well through small vessels in the body. Of
course, this means that organs and tissues supplied by these vessels become
starved for oxygen. This causes a very painful and debilitating condition
with significantly reduced life span.
Now,
there are several interesting observations to note. Most beneficial mutations
achieve their
benefits with just one or rarely two point mutations. Also, it is hard to
miss the fact that all of the functions gained, at least those listed here, were the result of a mutation that
interfered with a previously established function or specific interaction.
And, as we all know from a famous children's story, it is far easier to break than to create. The
reason is that there are so
many different ways to break something compared to the relatively few ways to make
something work. Why else couldn't all the King's men put Humpty Dumpty back together
again? (Back to Top)
Enzyme
Evolution
But,
how did such apparently complex enzymes, such as penicillinase, evolve? A
bacterium is not going to evolve the enzymatic penicillinase function with just
one or two point mutations to some target sequence because the penicillinase
function is not based on the loss or hindrance of a pre-existing function or
interaction. So, how could such a function evolve?
There are many
theories as to how the penicillinase enzyme must have evolved. However, when it comes
right down to it, no one has ever demonstrated the evolution of the
penicillinase enzyme in the lab. As previously noted, bacteria that
produce the penicillinase enzyme were always capable of producing this
enzyme or they obtained the code for this enzyme via a plasmid from another
bacterium who had this code already formed.11
Sometimes, a point mutation is required
to deregulate the production of penicillinase so that much greater quantities
are produced, rendering the bacterium (and its subsequence offspring) instantly
resistant to greater doses of penicillin. But, this change really has nothing to
do with explaining how the rather complex penicillinase function evolved.9
So, are there any documented reports of the evolution of a complex
enzymatic function comparable to that of the penicillinase function?
Michael
Behe, a professor of biochemistry at Lehigh University, says that,
“Molecular evolution is not based on scientific authority.
There is no publication in the scientific literature in prestigious
journals, specialty journals, or books that describe how molecular evolution of
any real, complex, biochemical system either did occur or even might have
occurred. There are assertions that
such evolution occurred, but absolutely none are supported by pertinent
experiments or calculations.†5
 Others,
such as the well known evolutionary biologist Kenneth Miller, disagree. In
his 1999 book, Finding Darwin’s God,
one of Miller’s challenges of Behe’s position includes a 1982 research study
by professor Barry Hall, an evolutionary biologist from the University of
Rochester. In this study, Hall deleted a gene (lacZ gene) in a type of
bacteria (E. coli) that makes an
enzyme (ß-galactosidase).
This enzyme converts the sugar lactose into the sugars glucose and
galactose. The E.
coli then use glucose and galactose for energy.
Others,
such as the well known evolutionary biologist Kenneth Miller, disagree. In
his 1999 book, Finding Darwin’s God,
one of Miller’s challenges of Behe’s position includes a 1982 research study
by professor Barry Hall, an evolutionary biologist from the University of
Rochester. In this study, Hall deleted a gene (lacZ gene) in a type of
bacteria (E. coli) that makes an
enzyme (ß-galactosidase).
This enzyme converts the sugar lactose into the sugars glucose and
galactose. The E.
coli then use glucose and galactose for energy.
Without
this lactase enzyme one would think that these bacteria and their offspring
would not be able to utilize lactose. However,
what Hall found is that after a short time (just one generation) of exposure to
a lactose-enriched environment these bacteria modified a different gene with just one
point mutation so that it gained the ability to produce a new
lactase enzyme.12 Since the original enzyme was composed of a
fairly large tetramer (~1,000 amino acids for each subunit), it seemed like the
evolution of the lactase function might require a fair amount of enzymatic
complexity (fairly large number and specific arrangement of amino acid
residues). In other words, it might be rather difficult to come across
very many enzymes with lactase ability out of the vast numbers of potential
arrangments within sequence space. So, the demonstration of such
rapid evolution of a completely different hexametric lactase enzyme was quite a
stunning success for Hall. How did these amazing bacteria evolve a brand
new enzyme to do such an apparently complex task?
As
it turns out, these E. coli bacteria had something of a spare tire gene
that Hall called the "evolved ß-galactosidase gene" (ebgA).
Just one point mutation was all it took to give this spare tire gene the ability
to produce a protein with the beneficial lactase activity. Hall was of
course disappointed to find out that only one point mutation was enough to
"evolve" this beneficial lactase activity. So, he did a very
interesting thing. He deleted both the original lacZ genes as well as the
evolved ebgA gene in some rather large colonies of E. coli. Interestingly
enough, none of these doubly mutated bacteria nor their offspring never evolved any
other gene or DNA sequence into a functional lactase enzyme despite observation
for tens of thousands of generations.
Hall was mystified. He described
these bacteria as having, "limited evolutionary potential." 12
The interesting thing is that these same bacteria that were limited in their
ability to evolve the lactase function would easily have evolved resistance to
just about any antibiotic in just a few generations of sublethal exposure. Compared to antibiotic
resistance, the evolution of even a single-protein enzyme is quite a
different matter. We are starting to climb the ladder of increasing
functional complexity. (Back
to Top)
Limited
Evolutionary Potential
Now
I ask, what exactly was limiting the evolutionary potential of Hall's bacteria?
Does the theory of evolution explain such limits? If so, how are they
explained? The theory of evolution claims the power to create incredible
diversity via mindless processes if given enough time. Well, how much
time, on average, would it take for E. coli, without lacZ and ebgA, to
evolve the lactase function? Can this time be estimated, even roughly?
If so, upon what basis can it be estimated?
According
to Hall's own calculations, a function that required just two independent
(neutral) mutations would take around "100,000 years" to achieve in E.
coli.12 It seems as though Hall does not understand the
statistics of random walk very well or he would not have been surprised when he
did in fact isolate such a double mutant in just a few days. The estimated
time for fixation is what caused Hall to estimate a time of 100,000 years for
the crossing of a gap of just two neutral mutations. What Hall did not
realize is that stepwise fixation of each mutation (spread to all members of a
population) is not required for such a gap to be crossed. With populations
the size of Hall's E. coli colonies, such a double mutation would be
realized in at least one bacterium in the population in just one or two generations using random walk alone. (Back
to Top)
Growing
Neutral Gaps
So,
is the problem solved? Hardly. With each doubling of the neutral gap
between the current genetic real estate and a new potentially beneficial
function, the random walk increases by a factor of two. For example, a gap of 2
amino acid residue differences has only 400 different options to fill (20
potentially different residues in each position in a protein sequence). A population of one
billion bacteria would quickly distribute itself among all these 400 options in
very short order (given that these 400 options were all functionally neutral
with respect to each other). However, doubling the gap to 4 differences would
increase the number of options to 160,000. Doubling the gap again
to 8 differences would increase the number of options to 25.6 billion. A gap
of 16 would yield 655 million trillion (6.5e20). In such a case, each
bacterium in the population of one billion would be surrounded by a sea
of 655 billion non-beneficial options. The time required to traverse this gap, even for a population of one billion bacteria, would run into the trillions
of generations. Why? Because the time required for a mutation to hit
even one of the residues that form the gap runs into the hundreds of thousands
of generations, on average. In other words, each random walk step would
take hundreds of thousands of generations. So, finding one specific spot
out of 655 billion options would require 6.55e9 x 1e5 = ~ 1e14 or 100 trillion
generations.
This is because natural selection cannot
preferentially select for any sequence that doesn't provide an improved function
over what was already there to begin with. The only forces for change that can sort
through such non-beneficial sequences are random mutations. These random
mutations randomly search through sequence space with the use of either short or
long random steps. However, regardless of the size of the step/mutation,
the odds of success are not changed. Such a random walk takes a whole lot more time than a
non-random direct walk would take - exponentially more time. This simple little problem is what
messes things all up for evolution.
For
instance, consider that there are many bacterial functions that are far more
complex than the relatively simple single-protein based enzymatic-type functions
of lactase or nylonase. Many single protein enzymes are actually fairly
complex, don't get me wrong. They certainly are far more complex than the
function of antibiotic resistance that arises via an interfering mutation.
However,
their functions are still relatively simple when compared to other cellular
functions of higher complexity.
For example, there are about 10130
potential protein sequences 100 residues in length. Of these 10130 potential proteins, how many
would have a specific function? Well, it depends on the function. It
depends upon how specific the arrangement of residues needs to be. So, as
an example, lets pick one of the more specifically arranged functional protein
that requires about 100 residues to work - like cytochrome c. Some
scientists, like Yockey, have estimated anywhere between 1050 to 1090
different potential cytochrome c proteins exist in sequence space.15-17 To understand how
big these numbers are, consider that the total number of atoms in the visible
universe is only around 1080. So, one can see that 1090
different cytochrome c sequences is an absolutely huge number. The problem
is, this pile
of 1090 cytochrome c proteins is absolutely miniscule when compared to 10130,
which is the total number of different potential protein sequences 100aa
in length. For every one cytochrome c sequence there would be about 1040
non-cytochrome c sequences in sequence space.
And
yet, this ratio gets exponentially worse as the complexity of function
increases. For example, the function of bacterial motility involves the
interactions of many different proteins all working together at the same time -
over 20 different structural protein parts in specific arrangement totaling well
over 10,000aa that must be specifically coded for in correct sequence.
The question is, how many different arrangements of these amino acid residues would
produce a motility system (or how many arrangements of 10,000 letters
would produce a meaningful, much less beneficial, essay in English)? For
argument's sake, lets say that 102000 different motility
systems could be made with such a stretch of amino acids. Despite this
apparently astronomical number of different motility systems, 102000
is still a tiny fraction of 1013,010 - the minimum potential protein
sequence space at this level of complexity (10,000aa level). Each sequence
with a motility function would be surrounded by at least 1011,000
sequences without the motility function. In fact, the beneficial sequence
density at this level of complexity seems to be so miniscule that evolution is
powerless to evolve any function at such a level of complexity. There
simply are no observable examples of any such function evolving in real time
- not one example (i.e., a function that requires more than a few thousand
fairly specifically arranged residues).
Now,
there are a whole lot of stories about how such functions must have evolved.
These stories are exclusively based on the notion that sequence similarities of
portions of such systems to portions of sequences in other systems of function
must mean that they share a common evolutionary ancestor. Certainly the
similarities do seem to indicate a common origin of some kind. However, as Behe
has repeatedly pointed out, nobody has provided any detailed explanation as to
how evolutionary mechanisms of random mutation and function-based selection
could give rise to the functional differences at higher levels of minimum
functional complexity. The functional differences are what are important
here - not so much the similarities. How are these differences explained
by any non-deliberate process? Both deliberate and non-deliberate
processes can explain the similarities, but how well can non-deliberate
evolutionary forces explain the differences?
The problem is that there is
always more potential junk than non-junk at any given level of complexity. The real
problem though is that this junk pile grows exponentially, relative to the pile
of potentially beneficial sequences, with each step up the
ladder of functional complexity. (Back
to Top)
The
Junk Pile and The Ladder of
Functional Complexity
The
ladder of complexity limits the ability of evolution to evolve beyond its lowest
rungs where the most simple functions, such as antibiotic resistance, can be
found. The target mutations required to achieve antibiotic resistance are
extremely simple to get "right" because there are so many
"right" options. However, the evolution of specific enzymatic
functions, like the lactase function, are a lot harder to get "right"
because far fewer options are "right." Then again, even these
functions are relatively easy to get "right" compared to more complex
multipart functions, such as bacterial motility systems, where all the protein parts
work together at the same time in a specific 3-D orientation with each other.
So, as one moves up the ladder of functional complexity, the difficulty of
finding any sequence that does anything beneficial at such a level of complexity
becomes exponentially harder and harder to do until not even trillions upon
trillions of years are enough. (Back
to Top)
A
Construction Foreman Who Cannot Read
 It
is all very much like a construction foreman who never learned how to read a
blueprint, but who intuitively knows what works when he sees it in action.
His workers are the ones that know how to read blueprints and follow
directions exactly. The workers
also copy parent blueprints to use as templates for each new house that is to be
built. However, although they are
extremely careful copyists, the workers make little mistakes every now an then.
These little mistakes may not result in any phenotypic change whatsoever,
but sometimes they translate into slight or even major variations among the
actual houses built (the phenotype). The
foreman then comes to inspect the completed houses and picks the one that is the
“best†given the particular needs of that house for that location and housing market.
The choice of the foreman is based only on current function.5,6
He knows only what works right now.
He has no imagination, memory, or vision for the future.
If there is a part of a house that he does not recognize as having
current beneficial overall function, he will not select to keep that house and
the offending part will be lost from future blueprint options. The foreman goes around saying,
“Keeping do-dads around that don’t work is expensive!â€Â
He will not maintain what he does not recognize as beneficial right now in hopes that sometime in
the future, with some potential change in the housing market/environment, it may
develop into something beneficial. Once
the selection for the best overall house is made by the foreman, the workers find the blueprint for this
house and use it as a template for the next building project.
It
is all very much like a construction foreman who never learned how to read a
blueprint, but who intuitively knows what works when he sees it in action.
His workers are the ones that know how to read blueprints and follow
directions exactly. The workers
also copy parent blueprints to use as templates for each new house that is to be
built. However, although they are
extremely careful copyists, the workers make little mistakes every now an then.
These little mistakes may not result in any phenotypic change whatsoever,
but sometimes they translate into slight or even major variations among the
actual houses built (the phenotype). The
foreman then comes to inspect the completed houses and picks the one that is the
“best†given the particular needs of that house for that location and housing market.
The choice of the foreman is based only on current function.5,6
He knows only what works right now.
He has no imagination, memory, or vision for the future.
If there is a part of a house that he does not recognize as having
current beneficial overall function, he will not select to keep that house and
the offending part will be lost from future blueprint options. The foreman goes around saying,
“Keeping do-dads around that don’t work is expensive!â€Â
He will not maintain what he does not recognize as beneficial right now in hopes that sometime in
the future, with some potential change in the housing market/environment, it may
develop into something beneficial. Once
the selection for the best overall house is made by the foreman, the workers find the blueprint for this
house and use it as a template for the next building project.
But
what happens if the housing market changes the next year?
What if certain changes would benefit the house in this new environment?
For example, what if there were a prolonged drought and wild fires became
a threat making houses with tile shingles more resistant to fire than houses
with wooden or asphalt shingles? Would
the foreman be able to make these beneficial changes?
Consider
the thought that languages and thus blueprints are arbitrary in that they use
arbitrary symbols to represent ideas. A
change or evolution of a symbol does not necessarily correlate to an equivalent
change in the attached idea. If a
symbol changes, even a little bit, the attached idea may simply disappear,
leaving the “new†symbol without any recognized function.
The symbol is now meaningless. For
example, what if the blueprint for our house in question called for “wooden
shingles.†Each of the words,
“wooden†and “shingles†is an arbitrary group of symbols that represents
an idea to an English speaking person. If
the blueprint could be changed to read “tile shinglesâ€Â, the understood
change in meaning and the resultant change in house building would be a clear
advantage in our fire-hazard environment. If
no pre-established alleles for “tile shingles†are available in the
blueprint pool of trait options, is there any way to create the “tile
allele†from anything that already exists in the gene pool?
The
problem for gradual change is that each letter change must make sense.
If “wood†is changed to “hoodâ€Â, the actual word “hood†has
meaning. However, is the meaning
for “hood†any closer to the meaning for “tile� Even
though hood has meaning, does it have beneficial meaning in this case? What
does “hood shingles†mean?
So,
not only does each word of the blueprint have to make sense to the workers, but
the combination or location of the words on the blueprint has to make sense as
well or else the workers cannot make anything, much less something
beneficial. Order is important at all
levels of complexity. Amino acid residue order is important for the proper
function of a single protein. Also, the order of multiple proteins is
important for the formation of a multiprotein system. Again, if the
workers build something that the foreman cannot recognize as beneficial right
now, it will be rejected. It is as simple as that.
In
order to better visualize the problem, put yourself in place of the foreman.
You can only select based on functions that work “right now.â€Â
With this in mind, consider the phrase, “Methinks it is like a
weasel.†6 Now, add,
subtract, or change one letter at a time from the phrase in any position or
order that you want. There are just
two more little rules to this game. Each
change that you make must make sense in English and each change must be
beneficial in a particular situation/environment.
See how far you can go and how much change in meaning you can get.
Changing the meaning very far is a lot more difficult than one might
think even if the beneficial nature of the change is not a concern.
Nature
runs into this same little problem. Changing genetic sequences too much destroys
all phenotypic function before any new function can be reached.
Maintaining a functional phenotypic trait along the path towards any
uniquely functional trait requires multiple changes (maybe even hundreds or
thousands at higher levels of functional complexity) that do not change the original function and do not achieve new
function, until all the changes are in place.5,12
This is because many functional genetic elements are isolated from each
other like islands on a large ocean of neutral function or even detrimental
function.
Consider the fact that most
character sequences of a given length have no
meaning to an English speaking person. The
same is true for sequences of DNA. The
vast majority of potential genes of a given length mean nothing to a given cell. Any
gene that happens to get mutated into one of these unrecognized or neutral genes
becomes suddenly lost in the ocean of neutrality where no guidance is
available. Without guidance,
evolution drowns in this ocean. (Back to Top)
Adding
a Stereo System to a Car
Some
say that evolution need not work like this but that the mindless processes of
nature evolve new traits and gene pools by simply adding on previously defined
genetic elements to an established system of function... like the addition of a
stereo system to a car. The addition of these units enhances the function
of the established system, even to the point of giving it new functions that it
never had before. In this way, a simple system of function can be enhanced
in complexity step by tiny step.
Let's
try a thought experiment to illustrate this point. Consider the sentence
in a large book of sentences that reads, "I am." Now add any
other word onto this sentence. The only rule is that whatever word you
choose must make sense in the context of the other sentences and the book as a
whole. For example, I could add the pre-formed word, "pleased"
and make the new sentence read, "I am pleased." This makes sense
in English and it adds meaning to the sentence (whether or not it makes sense to
the rest of the paragraph is another story). The new word,
"pleased" could have been the result of a duplication mutation of a
gene somewhere else that just happened to get inserted into our sentence.
But, if the mutation had read, "I am very", this phrase does not
necessarily make sense
in most situations/environments. The addition of the word "very"
probably destroyed the
previous function of our sentence without creating a new function.
However, if the phrase "I am pleased" was first to evolve, the phrase,
"I am very pleased" could evolve next and make sense.
With these
rules in mind, try and keep adding on words (or subtracting words) and see how
far you can get given a particular situation/environment. Maybe the next mutation could read, "I am very
pleased Tom." Then, "I am pleased Tom." Then, "I am
Tom." We could also go another route and say, "I am
very pleased in Tom." Then, "I am very pleased in seeing
Tom." Then "I am very pleased in seeing Tom run."
Then, "I am very pleased in seeing Tom run fast." Then, "I
am very pleased in seeing Tom run real fast." Then, etc. etc. etc.
We can evolve quite a few different phrases with quite a few unique meanings
with the simple addition or subtraction of previously defined words. Could
genes in DNA do the same thing? Technically yes, but there are just a few
problems to consider.
Remember
that not every defined word that exists in English will make sense when added to
the phrase, "I am." Granted, the odds that one will make
sense seem to be fairly good though. However, the longer our sentence
gets, the less words there are that make sense when they are added to our
sentence in the context of a specific situation/environment. Consider also that the placement of the words within our
sentence is extremely vital to the functionality of the sentence. I might
say, "I am green." This phrase makes sense in English. But
what if the word "green" had been inserted into the wrong place?
The sentence could just as easily have "evolved" to read, "I
agreenm." This makes no sense in English and destroys the function of
a previously functional sentence. Consider also that the duplication
mutation could have occurred or been inserted into an area of the book of
sentences where it was not needed. What are the odds that it would land in
exactly the right "evolving" sentence in exactly the right position
within that sentence when there are potentially millions of other locations it
could have landed? Then consider that the sentence itself, even if it
might make sense by itself, must make sense as it relates with the other
sentences around it and in the rest of the book. If any of these problems
arise, that sentence is lost in the ocean of non-beneficial function. (Back
to Top)
One
Last Question
If
the theory of
evolution runs into such apparently difficult statistical problems, how is it that
this theory can be so earnestly presented as the only
"rational" answer to the question of the origins of living things? (Back
to Top)
-
Lewin,
Benjamin, Genes V, Oxford
University Press, 1994.
-
Gelehrter,
Thomas D. et al. Principles of
Medical Genetics, 1998.
-
Callender,
L. A., Gregor Mendel: An opponent of descent with modification. History
of Science 26: 41-75. 1988.
-
Genetics
131: 245-253, 1992.
-
Behe,
Michael J. Darwin’s Black Box,
The Free Press, 1996.
-
Dawkins,
Richard. The Blind Watchmaker,
1987.
-
Mendel,
Gregor. Experiments in Plant Hybridization. 1865.
-
Veith,
Walter J. The
Genesis Conflict, The Amazing Discoveries Foundation, 1997, p.
82.
-
Carl
Wieland, Antibiotic Resistance in
Bacteria, Creation Ex Nihilo Technical Journal, 8:1 (1994), p. 5.
-
Felix
Konotey-Ahulu, The Sickle Cell Disease Patient (New York: Macmillan, 1991),
p. 106-108.
-
R.
McQuire, Eerie: human Arctic fossils
yield resistant bacteria, Medical Tribune, 12/29/1988, pp. 1, 23.
-
B.G.
Hall, Evolution on a Petri Dish. The
Evolved B-Galactosidase System as a Model for Studying Acquisitive Evolution
in the Laboratory, Evolutionary Biology, 15(1982): 85-150.
-
Dugaiczyk,
Achillies. Lecture Notes,
Biochemistry 110-A, University California Riverside, Fall 1999.
-
Ayala,
Francisco J.
Teleological Explanations in
Evolutionary Biology, Philosophy of Science, March, 1970, p. 3.
-
Yockey,
H.P., J Theor Biol, p. 91, 1981
-
Yockey,
H.P., "Information Theory and Molecular Biology", Cambridge
University Press, 1992
-
Sauer,
R.T. , James U Bowie, John F.R. Olson, and Wendall A. Lim, 1989,
'Proceedings of the National Academy of Science's USA 86, 2152-2156. and
1990, March 16, Science, 247; and, Olson and R.T. Sauer, 'Proteins:
Structure, Function and Genetics', 7:306 - 316, 1990.
(Back
to Top)
.
Home Page
. Truth,
the Scientific Method, and Evolution
.
Methinks
it is Like a Weasel
. The
Cat and the Hat - The Evolution of Code
.
Maquiziliducks
- The Language of Evolution
. Defining
Evolution
.
The
God of the Gaps
. Rube
Goldberg Machines
.
Evolving
the Irreducible
. Gregor
Mendel
.
Natural
Selection
. Computer
Evolution
.
The
Chicken or the Egg
. Antibiotic
Resistance
.
The
Immune System
. Pseudogenes
.
Genetic
Phylogeny
. Fossils
and DNA
.
DNA
Mutation Rates
. Donkeys,
Horses, Mules and Evolution
.
The
Fossil Record
. The
Geologic Column
.
Early Man
. The
Human Eye
.
Carbon
14 and Tree Ring Dating
. Radiometric
Dating
.
Amino Acid
Racemization Dating
. The
Steppingstone Problem
.
Quotes
from Scientists
. Ancient
Ice
.
Meaningful
Information
. The
Flagellum
.
Harlen Bretz
. Milankovitch
Cycles
Debates:
Stacking
the Deck
God
of the Gaps
The
Density of Beneficial Functions
All
Functions are Irreducibly Complex
Ladder
of Complexity
Chaos
and Complexity
Confusing
Chaos with Complexity
Evolving
Bacteria
Irreducible
Complexity
Scientific
Theory of Intelligent Design
A
Circle Within a Circle
Crop
Circles
Mindless
vs. Mindful
Single
Protein Functions
BCR/ABL
Chimeric Protein
Function
Flexibility
The
Limits of Functional Flexibility
Functions
based on Deregulation
Neandertal
DNA
Human/Chimp
phylogenies
Geology
The
Geologic Column
Fish
Fossils
Matters
of Faith
Evidence
of Things Unseen
The
Two Impossible Options
Links
to Design, Creation, and Evolution Websites
Since
June 1, 2002

 The
theory of evolution proposes that all the various blueprints of living things
are descendents of a single common ancestor blueprint.
The diversity of blueprints that exist today is simply the result of
variations on a single theme. If
true, the power of the mindless evolutionary force of nature is truly
astounding. The sheer creativity
and magnificent diversity of nature is enough to make even the most cynical
stand in silent awe. If humankind
could harness this power and speed it up with the aid of our intelligent minds,
the implications for advancement seem unlimited.
The
theory of evolution proposes that all the various blueprints of living things
are descendents of a single common ancestor blueprint.
The diversity of blueprints that exist today is simply the result of
variations on a single theme. If
true, the power of the mindless evolutionary force of nature is truly
astounding. The sheer creativity
and magnificent diversity of nature is enough to make even the most cynical
stand in silent awe. If humankind
could harness this power and speed it up with the aid of our intelligent minds,
the implications for advancement seem unlimited.  Darwin
was unable to answer this question although the answer was in fact available in
his own day.
Darwin
was unable to answer this question although the answer was in fact available in
his own day. The
variation in allelic expression can be quite dramatic.
The
variation in allelic expression can be quite dramatic. Others,
such as the well known evolutionary biologist Kenneth Miller, disagree. In
his 1999 book, Finding Darwin’s God,
one of Miller’s challenges of Behe’s position includes a 1982 research study
by professor Barry Hall, an evolutionary biologist from the University of
Rochester. In this study, Hall deleted a gene (lacZ gene) in a type of
bacteria (E. coli) that makes an
enzyme (
Others,
such as the well known evolutionary biologist Kenneth Miller, disagree. In
his 1999 book, Finding Darwin’s God,
one of Miller’s challenges of Behe’s position includes a 1982 research study
by professor Barry Hall, an evolutionary biologist from the University of
Rochester. In this study, Hall deleted a gene (lacZ gene) in a type of
bacteria (E. coli) that makes an
enzyme ( It
is all very much like a construction foreman who never learned how to read a
blueprint, but who intuitively knows what works when he sees it in action.
It
is all very much like a construction foreman who never learned how to read a
blueprint, but who intuitively knows what works when he sees it in action.